Two Postdoc are leaving the lab for new adventures!
Yesterday we celebrated the departure of two talented postdocs, Luke Harrison and Alex Rivera-Millot, who are leaving for exciting new adventures — Luke as a Professor at McGill University in Montreal, and Alex as a Chargé de Recherche (CR) at the CNRS in France.
A big thank-you to both of them for their contributions, and best of luck in their future careers — I’m sure they will be nothing short of amazing!


A new tool for ssDNA technology!
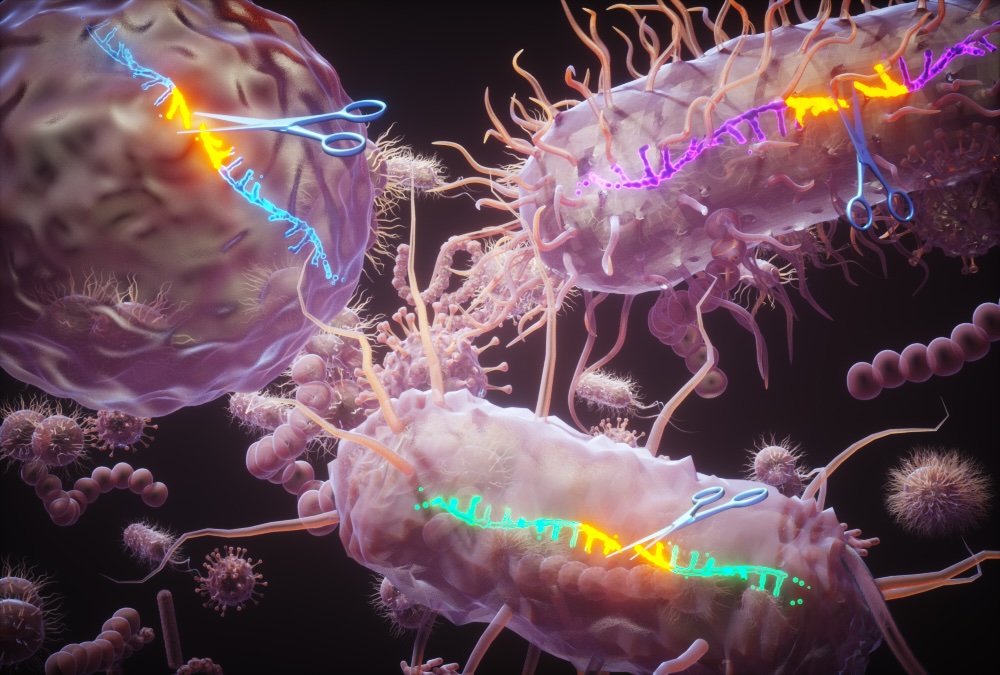
Fifteen years ago, while studying the manganese exporter MntX in Neisseria, I noticed some intriguing repeated sequences surrounding the gene. It took us all this time to fully understand their role and discover the factor that target them. In this Nature Communications article , we introduce a new family of proteins capable of cleaving a specific sequence in single-stranded DNA, similar to how restriction enzymes target double-stranded DNA. To our knowledge, this activity has not been previously described and opens the door to a wide range of applications involving ssDNA. We’ve filed a patent to continue exploring this promising discovery. We are now also searching for partners with ssDNA applications and problematics!
There are two main findings in this study: First, we describe a large family of sequence-specific single-stranded DNA (ssDNA) enzymes, now named Ssn, which are capable of cleaving their own specific sequence in ssDNA. Second, we uncover the role these sequences and Ssn proteins play in Neisseria, including the pathogens N. meningitidis and N. gonorrhoeae. We propose that these mechanisms may represent a novel « dialect » these species use to regulate gene exchange with other organisms. A lot more to come 🙂
CIHR Project Grant Funded!!!!!
Our project « Deciphering the stepwise cell envelope evolution that allowed Neisseria nasopharyngeal adaptation and pathogenesis. » has been funded by CIHR. A great news for the lab for the next five years!

A look back on 2020:
The year 2020 is almost finished (ouf). A crazy year with a worldwide pandemic that reminded us of the need for science and the importance of our work (even if it is a small piece). Despite this, the lab has some amazing good news, here is a sample of them:
January:
Our grant « Deciphering Neisseria pathogens cell envelope evolution: optimization for virulence or Achilles’ heel of N. meningitidis and N. gonorrhoeae? » has been partially funded by

March: We received funding to build on our imagery platform (electron microscopy and animal imaging) from:
May: Martin wrote an article on Nightlife about COVID-19 fake news:

June: Martin Chenal (Ph.d. in the lab) got the first place at the INRS contest « Ma thèse en 180s »
July: Our collaboration with Nick Doucet on the genome of Dictyopanus pusillus, the first eukaryote we have sequenced, is out:

August: Our amazing postdoc Antony Vincent got his professor appointment at Université Laval:

September: Our article on Sigma Factor regulation, in collaboration with Marcel Behr, is out:

October: Our scientific article about regulation of Sigma factor activity via methylation of promoters in Leptospira is out:
See press release:

November: 1) Our scientific article about a new compound that selectively kills Neisseria pathogens is out:

2) A press release on that topic and several articles in the media:
3) Eve Bernet (Ph.d. in the lab) received the first prize for her Poster presentation at the JQRSR
4) Our collaboration with Yves St-Pierre about Mycobacteria and mussels is out:

5) Martin Chenal (Ph.d.) in the lab got the second prize at the national final of « Ma thèse en 180s » (3 minute thesis)
6) 3 students from the lab (Ève Bernet, Martin Chenal, Juan Guerra) officially completed their M.Sc. and obtained their diploma. Two of them (Ève and Martin) are staying with us for a PhD.
December: I got my tenure associate prof promotion !!!! and Our article CAPRIB is out:

We are back!
After few months at home students are back in the lab… Check out soon their amazing work to be released in the next couple of days.
Thanks to my amazing 2020 team!
Felicitation Marthe Lebughe: INRS – Innovation price

Good Luck in your future Career. It was sometime difficult but you never quit and you made it to the end with honors. Thanks for your effort and your trust.


